Putting it all together: Running a GOMC Simulation
It is strongly recommended that you download the test system provided at GOMC Website or Our Github Page
Run different simulation types in order to become more familiar with different parameter and configuration files (*.conf).
To recap the previous examples, a simulation of isobutane will be completed for a single temperature point on the saturated vapor-liquid coexistence curve.
The general plan for running the simulation is:
Build GOMC (if not done already)
Copy GOMC executable to build directory
Create scripts, PDB, and topology file to build the system, plus in.dat file and parameter files to prepare for runtime
Build finished PDBs and PSFs using the simulation.
Run the simulation in the terminal.
Analyze the output.
Please, complete steps 1 and 2; then, traverse to the directory, which should now contain a single file “GOMC_CPU_GEMC”. Next, six files need to be made:
PDB file for isobutane
Topology file describing isobutane residue
Two
*.inppackmol scripts to pack two system boxesTwo TCL scripts to input into
PSFGento generate the final configuration
isobutane.pdb
REMARK 1 File created by GaussView 5.0.8
ATOM 1 C1 ISB 1 0.911 -0.313 0.000 C
ATOM 2 C2 ISB 1 1.424 -1.765 0.000 C
ATOM 3 C3 ISB 1 -0.629 -0.313 0.000 C
ATOM 4 C4 ISB 1 1.424 0.413 -1.257 C
END
Top_Branched_Alkane.inp
* Custom top file -- branched alkanes
*
MASS 1 CH3 15.035 C !
MASS 2 CH1 13.019 C !
AUTOGENERATE ANGLES DIHEDRALS
RESI ISB 0.00 ! isobutane { TraPPE }
GROUP
ATOM C1 CH1 0.00 ! C3\
ATOM C2 CH3 0.00 ! C2-C1
ATOM C3 CH3 0.00 ! C4/
ATOM C4 CH3 0.00 !
BOND C1 C2 C1 C3 C1 C4
PATCHING FIRS NONE LAST NONE
END
pack_box_0.inp
tolerance 3.0
filetype pdb
output STEP2_ISB_packed_BOX_0.pdb
structure isobutane.pdb
number 1000
inside cube 0. 0. 0. 68.00
end structure
pack_box_1.inp
tolerance 3.0
filetype pdb
output STEP2_ISB_packed_BOX_1.pdb
structure isobutane.pdb
number 1000
inside cube 0. 0. 0. 68.00
end structure
build_box_0.inp
package require psfgen
topology ./Top Branched Alkane.inp segment ISB {
pdb ./STEP2_ISB_packed_BOX_0.pdb
first none
last none
}
coordpdb ./STEP2 ISB_packed_BOX_0.pdb ISB
writepsf ./STEP3_START_ISB_sys_BOX_0.psf
writepdb ./STEP3_START_ISB_sys_BOX_0.pdb
build_box_1.inp
package require psfgen
topology ./Top Branched Alkane.inp segment ISB {
pdb ./STEP2_ISB_packed_BOX_1.pdb
first none
last none
}
coordpdb ./STEP2 ISB_packed_BOX_1.pdb ISB
writepsf ./STEP3_START_ISB_sys_BOX_1.psf
writepdb ./STEP3_START_ISB_sys_BOX_1.pdb
These files can be created with a standard Linux or Windows text editor. Please, also copy a Packmol executable into the working directory.
Once those files are created, run in the terminal:
$ ./packmol < pack_box_0.inp
$ ./packmol < pack_box_1.inp
This will create the intermediate PDBs.
Then, run the PSFGen scripts to finish the system using the following commands:
$ vmd -dispdev text < ./build_box_0.inp
$ vmd -dispdev text < ./build_box_1.inp
This will create the intermediate PDBs.
To run the code a few additional things will be needed:
A GOMC Gibbs ensemble executable
A control file
Parameter files
Enter the control file (in.conf) in the text editor in order to modify it. Example files for different simulation types can be found in previous section.
Once these four files have been added to the output directory, the simulation is ready.
Assuming the code is named GOMC_CPU_GEMC, run in the terminal using:
$ ./GOMC CPU GEMC in.conf > out_ISB_T_330.00_K_RUN_0.log &
For running GOMC in parallel, using openmp, run in the terminal using:
$ ./GOMC CPU GEMC +p4 in.conf > out_ISB_T_330.00_K_RUN_0.log&
Here, 4 defines the number of processors that will be used to run the simulation in parallel.
Progress can be monitored in the terminal with the tail command:
$ tail -f out_ISB.log
Attention
Congratulations! You have examined a single-phase coexistence point on the saturated vapor-liquid curve using GOMC operating in the Gibbs ensemble.
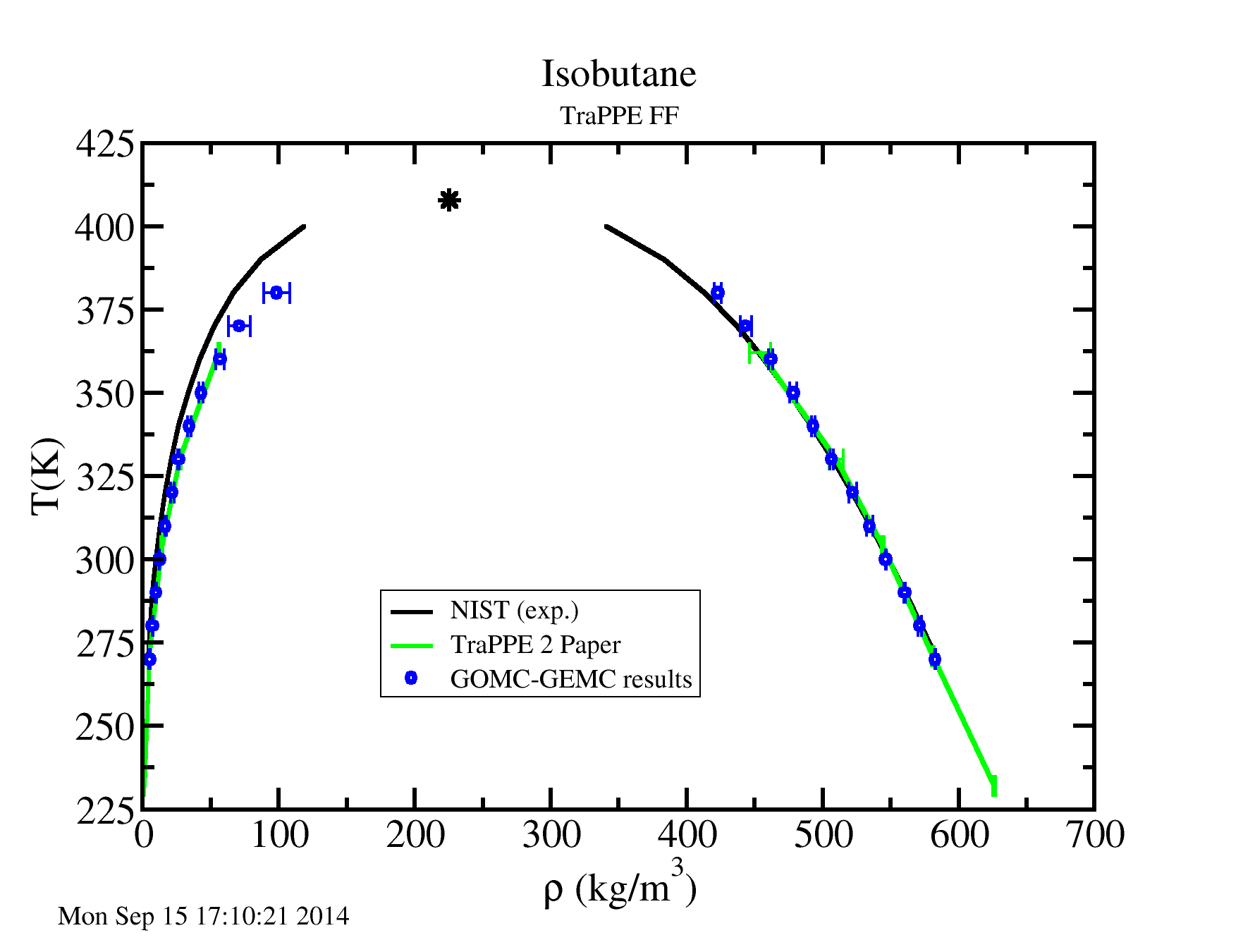
Repeating this process for multiple temperatures will allow you to obtain the following results.